Ping Wang' Lab @ HUST-Molecular Biology and Molecular Imaging
As a highly interdisciplinary research team, our graduate students came from the Department of Physics, Chemistry, Biology, EE, ECE, Optoelectronics and many other schools. We work passionately for the goal to explore and innovate molecular imaging techniques in basic life science and clinical applications. By pursuing quantitative and precision measurements in biology as it is in physics, our enthusiasm focuses on optical super-resolution imaging, deep tissue imaging, tissue tomography, smart genetic Raman tags to overcome the current instrumental limitations and answer important biological questions through our new ideas and even sudden inspiration after years of pondering. These new and robust methods will substantially extend our capabilities in observation of the nano-architectures of single cells and help us access the genetic trees of proteins and metabolites via molecular Raman imaging. Life is based on chemicals and unthinkable high-efficient chemical reactions. We set our mind on contributing our faint lives and energy to developing magic optical imaging tools to help us look into the life more closely. Hopefully, it can be the most disruptive one in the future. (Ping Wang July 2020)
Huazhong University of Science and Technology @ Wuhan 武汉光电国家研究中心
Research Interests
Label-free Hyperspectral Stimulated Raman Scattering Microscopy
Lipid metabolism is dysregulated in human cancers. The analytical tools that could identify and quantitatively map metabolites in unprocessed human tissues with submicrometer resolution are highly desired. Here, we implemented analytical hyperspectral stimulated Raman scattering microscopy to map the lipid metabolites in situ in normal and cancerous liver tissues from 24 patients. In contrast to the conventional wisdom that unsaturated lipid accumulation enhances tumor cell survival and proliferation, we unexpectedly visualized substantial amount of saturated fat accumulated in cancerous liver tissues, which was not seen in majority of their adjacent normal tissues. Further analysis by mass spectrometry confirmed significant high levels of glyceryl tripalmitate specifically in cancerous liver. These findings suggest that the aberrantly accumulated saturated fat may have great potential to be a metabolic biomarker for liver cancer.
1. S. Yan#, S. S. Cui #, K. Ke, B. X. Zhao, X. L. Liu, S. H. Yue*, P. Wang*. Hyperspectral Stimulated Raman Scattering Microscopy Unravels Aberrant Accumulation of Saturated Fat in Human Liver Cancer . Anal. Chem. 90 , 6362-6366 (2018)
2. B. Huang#, S. Yan#, L. Xiao, R. Ji, L. Yang, A. J. Miao*, P. Wang*. Label-free Imaging of Nanoparticle Uptake Competition in Single Cells by Hyperspectral Stimulation Raman Scattering . Small 14 (2018)
-
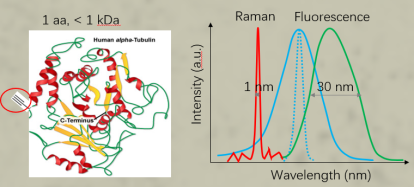
UAA Carried Raman Tag for Genetically Targeted Protein Imaging
Raman has been implemented to image biological systems for decades. However, Raman microscopy along with Raman probes is restricted to image metabolites or a few intracellular organelles so far and lacks genetic specificity for imaging proteins of interest, which significantly hinders their application. Here, we report the Raman spectra-based protein imaging method, which incorporates a small phenyl ring enhanced Raman tag (total of ∼0.55 kDa) with a single unnatural amino acid (UAA) to genetically label specific proteins. We further demonstrate hyperspectral stimulated Raman scattering (SRS) imaging of the Histone3.3 protein in the nucleus, Sec61β protein in the endoplasmic reticulum of HeLa cells, and Huntingtin protein Htt74Q in mutant huntingtin-induced cells. Genetic encoding of a small, stable, sensitive, and narrow-band Raman tag took one key step forward to enable SRS or Raman imaging of specific proteins and could further facilitate quantitative Raman spectra-based supermultiplexing microscopy in the future.
1. J. Zhang#, S. Yan#, Z. He, C. Ding, T. Zhai, Y. Chen, H. Li, G. Yang, X. Zhou, P. Wang*. Small Unnatural Amino Acid Carried Raman Tag for Molecular Imaging of Genetically Targeted Proteins. The journal of physical chemistry letters 9, 4679-4685 (2018)
2. S. Tian#, H. Z. Li#, Z. Li#, H. J. Tang, M. M. Yin, Y. G. Chen, S. Wang, Y. T. Gao, X. L. Yang, F. L. Meng*, J. W. Lauher*, P. Wang*, L.Luo*. Polydiacetylene-based Ultrastrong Bioorthogonal Raman Probes for Targeted Live-cell Raman Imaging. Nature Communications 11, 81 (2020)
High-resolution Stimulated Raman Scattering Microscope beyond 130 nm
High-resolution optical microscopes that can break 180 nm in spatial resolution set to conventional microscopies are much-needed tools. However, current optical microscopes have to rely on exogenous fluorescent labels to achieve high resolution in biological imaging. Herein, we report near-resonance enhanced label-free stimulated Raman scattering (SRS) microscopy with a lateral resolution near 130 nm, in which the high-resolution image contrast originates directly from a low concentration of endogenous biomolecules, with sensitivity gains of approximately 23 times. Moreover, by using a 0.3-m-long optical fiber, we developed hyperspectral SRS microscopy based on spectral focusing technology. Attributed to enhancements in spatial resolution and sensitivity, we demonstrated high-resolution imaging of three-dimensional structures in single cells and high-resolution mapping of large-scale intact mouse brain tissues in situ. By using enhanced high-resolution hyperspectral SRS, we chemically observed sphingomyelin distributed in the myelin sheath that insulates single axons. Our concept opens the door to biomedical imaging with ~130 nm resolution.
Y. Bi#, C. Yang#, Y. Chen, S. Yan, G. Yang, Y. Wu, G. Zhang*, P. Wang*. Near-resonance enhanced label-free stimulated Raman scattering microscopy with spatial resolution near 130 nm. Light: science & applications 7, 81 (2018)
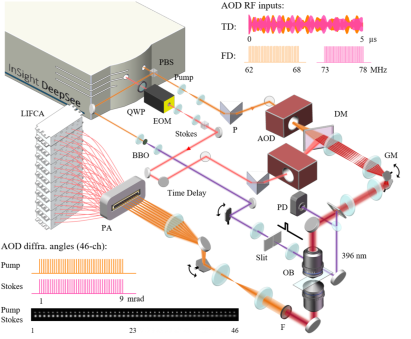
2 Khz Ultrafast SRS Microscope
Numerous mechanisms have been proposed for polymerization to provide qualitative and quantitative prediction of how monomers spatially and temporally arrange into the polymeric chains. However, less is known about this process at the molecular level because the ultrafast chemical reaction is inaccessible for any form of microscope so far. Here, to address this unmet challenge, a stimulated Raman scattering microscope based on collinear multiple beams (COMB‐SRS) is demonstrated, which allows label‐free molecular imaging of polymer synthesis in action at speed of 2000 frames per second. The field of view of the developed 2 kHz SRS microscope is 30 × 28 µm2 with 50 × 46 pixels and 7 µs dwell time. By catching up the speed of chemical reaction, COMB‐SRS is able to quantitatively visualize the ultrafast dynamics of molecular vibrations with submicron spatial resolution and sub‐millisecond temporal resolution. The propagating polymer waves driven by reaction rate and persistent UV initiation are observed in situ. This methodology is expected to permit the development of novel functional polymers, controllable photoresists, 3D printing, and other new polymerization technologies.
H. Li#, Y. Cheng, H. Tang, Y. Bi, Y. Chen, G. Yang, S. Guo, S. Tian, J. Liao, X. Lv, S. Zeng, M. Zhu, C. Xu, J. Cheng, P. Wang*. Imaging Chemical Kinetics of Radical Polymerization with an Ultrafast Raman Microscope. Advanced Science 7, 1903644 (2020)